El genotipo de un organismo es su conjunto completo de material genético. [1] El genotipo también puede usarse para referirse a los alelos o variantes que un individuo porta en un gen o ubicación genética en particular. [2] La cantidad de alelos que un individuo puede tener en un gen específico depende de la cantidad de copias de cada cromosoma que se encuentran en esa especie, también conocida como ploidía . En especies diploides como los humanos, están presentes dos juegos completos de cromosomas, lo que significa que cada individuo tiene dos alelos para cualquier gen dado. Si ambos alelos son iguales, el genotipo se denomina homocigoto . Si los alelos son diferentes, el genotipo se denomina heterocigoto.
El genotipo contribuye al fenotipo , los rasgos y características observables en un individuo u organismo. [3] El grado en que el genotipo afecta al fenotipo depende del rasgo. Por ejemplo, el color de los pétalos en una planta de guisantes está determinado exclusivamente por el genotipo. Los pétalos pueden ser morados o blancos dependiendo de los alelos presentes en la planta de guisantes. [4] Sin embargo, otros rasgos solo están parcialmente influenciados por el genotipo. Estos rasgos a menudo se denominan rasgos complejos porque están influenciados por factores adicionales, como factores ambientales y epigenéticos . No todos los individuos con el mismo genotipo se ven o actúan de la misma manera porque la apariencia y el comportamiento se modifican por las condiciones ambientales y de crecimiento. Del mismo modo, no todos los organismos que se parecen necesariamente tienen el mismo genotipo.
El término genotipo fue acuñado por el botánico danés Wilhelm Johannsen en 1903. [5]
Cualquier gen dado suele provocar un cambio observable en un organismo, conocido como fenotipo. Los términos genotipo y fenotipo son distintos por al menos dos razones:
A simple example to illustrate genotype as distinct from phenotype is the flower colour in pea plants (see Gregor Mendel). There are three available genotypes, PP (homozygous dominant), Pp (heterozygous), and pp (homozygous recessive). All three have different genotypes but the first two have the same phenotype (purple) as distinct from the third (white).
A more technical example to illustrate genotype is the single-nucleotide polymorphism or SNP. A SNP occurs when corresponding sequences of DNA from different individuals differ at one DNA base, for example where the sequence AAGCCTA changes to AAGCTTA.[6] This contains two alleles : C and T. SNPs typically have three genotypes, denoted generically AA Aa and aa. In the example above, the three genotypes would be CC, CT and TT. Other types of genetic marker, such as microsatellites, can have more than two alleles, and thus many different genotypes.
Penetrance is the proportion of individuals showing a specified genotype in their phenotype under a given set of environmental conditions.[7]
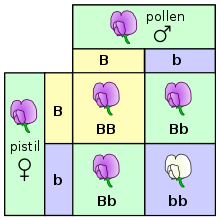
Traits that are determined exclusively by genotype are typically inherited in a Mendelian pattern. These laws of inheritance were described extensively by Gregor Mendel, who performed experiments with pea plants to determine how traits were passed on from generation to generation.[8] He studied phenotypes that were easily observed, such as plant height, petal color, or seed shape.[8] He was able to observe that if he crossed two true-breeding plants with distinct phenotypes, all the offspring would have the same phenotype. For example, when he crossed a tall plant with a short plant, all the resulting plants would be tall. However, when he self-fertilized the plants that resulted, about 1/4 of the second generation would be short. He concluded that some traits were dominant, such as tall height, and others were recessive, like short height. Though Mendel was not aware at the time, each phenotype he studied was controlled by a single gene with two alleles. In the case of plant height, one allele caused the plants to be tall, and the other caused plants to be short. When the tall allele was present, the plant would be tall, even if the plant was heterozygous. In order for the plant to be short, it had to be homozygous for the recessive allele.[8][9]
One way this can be illustrated is using a Punnett square. In a Punnett square, the genotypes of the parents are placed on the outside. An uppercase letter is typically used to represent the dominant allele, and a lowercase letter is used to represent the recessive allele. The possible genotypes of the offspring can then be determined by combining the parent genotypes.[10] In the example on the right, both parents are heterozygous, with a genotype of Bb. The offspring can inherit a dominant allele from each parent, making them homozygous with a genotype of BB. The offspring can inherit a dominant allele from one parent and a recessive allele from the other parent, making them heterozygous with a genotype of Bb. Finally, the offspring could inherit a recessive allele from each parent, making them homozygous with a genotype of bb. Plants with the BB and Bb genotypes will look the same, since the B allele is dominant. The plant with the bb genotype will have the recessive trait.
These inheritance patterns can also be applied to hereditary diseases or conditions in humans or animals.[11][12][13] Some conditions are inherited in an autosomal dominant pattern, meaning individuals with the condition typically have an affected parent as well. A classic pedigree for an autosomal dominant condition shows affected individuals in every generation.[11][12][13]

Other conditions are inherited in an autosomal recessive pattern, where affected individuals do not typically have an affected parent. Since each parent must have a copy of the recessive allele in order to have an affected offspring, the parents are referred to as carriers of the condition.[11][12][13]In autosomal conditions, the sex of the offspring does not play a role in their risk of being affected. In sex-linked conditions, the sex of the offspring affects their chances of having the condition. In humans, females inherit two X chromosomes, one from each parent, while males inherit an X chromosome from their mother and a Y chromosome from their father. X-linked dominant conditions can be distinguished from autosomal dominant conditions in pedigrees by the lack of transmission from fathers to sons, since affected fathers only pass their X chromosome to their daughters.[13][14][15] In X-linked recessive conditions, males are typically affected more commonly because they are hemizygous, with only one X chromosome. In females, the presence of a second X chromosome will prevent the condition from appearing. Females are therefore carriers of the condition and can pass the trait on to their sons.[13][14][15]
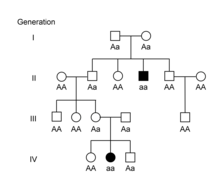
Mendelian patterns of inheritance can be complicated by additional factors. Some diseases show incomplete penetrance, meaning not all individuals with the disease-causing allele develop signs or symptoms of the disease.[13][16][17] Penetrance can also be age-dependent, meaning signs or symptoms of disease are not visible until later in life. For example, Huntington disease is an autosomal dominant condition, but up to 25% of individuals with the affected genotype will not develop symptoms until after age 50.[18] Another factor that can complicate Mendelian inheritance patterns is variable expressivity, in which individuals with the same genotype show different signs or symptoms of disease.[13][16][17] For example, individuals with polydactyly can have a variable number of extra digits.[16][17]
Many traits are not inherited in a Mendelian fashion, but have more complex patterns of inheritance.
For some traits, neither allele is completely dominant. Heterozygotes often have an appearance somewhere in between those of homozygotes.[19][20] For example, a cross between true-breeding red and white Mirabilis jalapa results in pink flowers.[20]
Codominance refers to traits in which both alleles are expressed in the offspring in approximately equal amounts.[21] A classic example is the ABO blood group system in humans, where both the A and B alleles are expressed when they are present. Individuals with the AB genotype have both A and B proteins expressed on their red blood cells.[21][22]
Epistasis is when the phenotype of one gene is affected by one or more other genes.[23] This is often through some sort of masking effect of one gene on the other.[24] For example, the "A" gene codes for hair color, a dominant "A" allele codes for brown hair, and a recessive "a" allele codes for blonde hair, but a separate "B" gene controls hair growth, and a recessive "b" allele causes baldness. If the individual has the BB or Bb genotype, then they produce hair and the hair color phenotype can be observed, but if the individual has a bb genotype, then the person is bald which masks the A gene entirely.
A polygenic trait is one whose phenotype is dependent on the additive effects of multiple genes. The contributions of each of these genes are typically small and add up to a final phenotype with a large amount of variation. A well studied example of this is the number of sensory bristles on a fly.[25] These types of additive effects is also the explanation for the amount of variation in human eye color.
Genotyping refers to the method used to determine an individual's genotype. There are a variety of techniques that can be used to assess genotype. The genotyping method typically depends on what information is being sought. Many techniques initially require amplification of the DNA sample, which is commonly done using PCR.
Algunas técnicas están diseñadas para investigar SNP o alelos específicos en un gen o conjunto de genes en particular, como si un individuo es portador de una condición particular. Esto se puede hacer a través de una variedad de técnicas, incluidas las sondas de oligonucleótidos específicos de alelo (ASO) o la secuenciación de ADN . [26] [27] También se pueden utilizar herramientas como la amplificación de sonda dependiente de ligación múltiple para buscar duplicaciones o deleciones de genes o secciones de genes. [27] Otras técnicas están destinadas a evaluar una gran cantidad de SNP en todo el genoma, como las matrices de SNP . [26] [27] Este tipo de tecnología se utiliza comúnmente para estudios de asociación de todo el genoma .
También se dispone de técnicas a gran escala para evaluar el genoma completo. Esto incluye el cariotipo para determinar la cantidad de cromosomas que tiene un individuo y microarreglos cromosómicos para evaluar grandes duplicaciones o deleciones en el cromosoma. [26] [27] Se puede determinar información más detallada utilizando la secuenciación del exoma , que proporciona la secuencia específica de todo el ADN en la región codificante del genoma, o la secuenciación del genoma completo , que secuencia todo el genoma, incluidas las regiones no codificantes. [26] [27]
En los modelos lineales, los genotipos pueden codificarse de diferentes maneras. Consideremos un locus bialélico con dos alelos posibles, codificados por y . Consideramos que corresponde al alelo dominante el alelo de referencia . La siguiente tabla detalla las diferentes codificaciones. [28]
{{cite book}}: CS1 maint: location missing publisher (link) which was rewritten, enlarged and translated into German as Johannsen W (1905). Elemente der exakten Erblichkeitslehre (in German). Jena: Gustav Fischer. Archived from the original on 2009-05-30. Retrieved 2017-07-19.